Abstract Authors
Darin Edward Holman - Environmental Biotechnology Laboratory, Department of Biotechnology, University of the Western Cape
Ashwil Klein - Plant Omics Laboratory, Department of Biotechnology, University of the Western Cape
Marshall Keyster - Environmental Biotechnology Laboratory, Department of Biotechnology, University of the Western Cape
Abstract Description
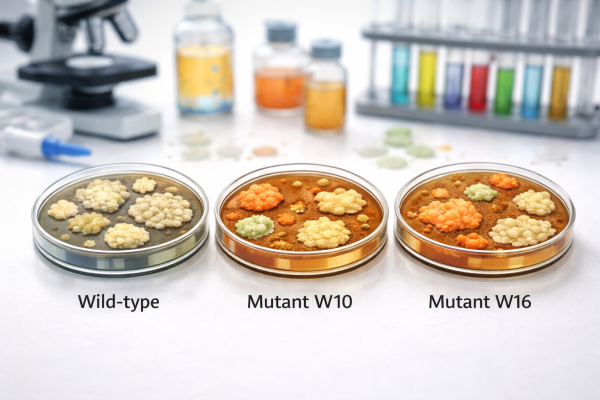
This study investigates the pathogenicity mechanisms of Pantoea agglomerans R6, a bacterial strain isolated from the root tissue of Lactuca serriola, with implications for both plant and human health. Using ethyl methanesulfonate (EMS) mutagenesis, we aimed to identify genetic and/or metabolic factors contributing to the pathogenicity of this strain. Mutations were successfully induced in P. agglomerans R6 using EMS concentrations of 1.25 mM and 2.5 mM with incubation periods of 30 and 60 minutes. These conditions generated mutant strains displaying notable phenotypic changes, including altered colony morphology and biochemical activity. Two mutants, W10 and W16, were selected based on their ability to increase the germination percentage of Brassica napus seeds.
Comprehensive characterization of these mutants, alongside the wild-type (WT-R6) strain, included biochemical, genomic, and metabolomic analyses. Sequencing of the 16S rDNA region revealed sequence variation between WT-R6 and the mutants. However, as the 16S rDNA gene encodes ribosomal RNA rather than a protein, mutations in this region do not result in truncated protein products and should not be interpreted in terms of premature stop codons affecting translation. In addition, the interpretation of 16S rDNA sequence differences was considered in light of (i) the redundancy of 16S rRNA gene copies in many bacterial genomes and (ii) the potential influence of primer selection and PCR amplification bias on the recovery of variant 16S sequences, particularly when the observation was not derived from whole genome sequencing. Phylogenetic analysis indicated that W10 and W16 are closely related to each other, while showing divergence from WT-R6 based on the amplified 16S rDNA sequence data.
Whole genome sequencing of WT-R6 provided broader genetic insights, including protein-coding genes, RNA genes, pseudogenes, and GC content. Importantly, genome annotation identified putative virulence factors linked to secretion systems, adhesion, motility, iron uptake, and toxin-associated functions, suggesting potential mechanisms by which WT-R6 may interact with host tissues and contribute to disease development. Metabolomic profiling further revealed that WT-R6 produces distinct metabolites, including diindolylmethane and indoleacetic acid, which may contribute to host interaction. A secondary metabolite of interest, Polanrazine B, was also detected and is noteworthy given its reported association with pathogenicity in Leptosphaeria maculans affecting B. napus. In addition, distinct volatile organic compound (VOC) profiles were observed among WT-R6 and its mutants, supporting the possibility that volatile-mediated signaling contributes to bacterial-host communication.
Overall, this study provides insights into the genetic and metabolic features associated with P. agglomerans R6 pathogenicity. The identification of virulence-associated traits such as secretion systems, motility mechanisms, and toxin-related functions, together with candidate pathogenic metabolites (including Polanrazine B potentially linked to an NRPS biosynthetic gene cluster), lays the groundwork for future work aimed at mitigating bacterial pathogenicity and supporting sustainable agricultural applications, while also informing broader antimicrobial research.

